
Beamforming with ADMIRE (Aperture Domain Model Image REconstruction) and Deep Networks

Our current beamforming efforts focus on non-linear beamforming and filtering strategies for aperture domain signals. The aperture domain signal (channel data) is the RF signal sampled by each transducer element. The transducer channel data contains an amazing amount of information. Extracting this information has been a question for decades in many fields including radar, sonar, and medical ultrasound. One challenge of extracting the information is having an adequate expectation of the information that will be present (a model). A significant feature is multiple scattering. Multipath scattering is usually ignored in ultrasound, but the movie on the right (courtesy of Jeremy Dahl at Stanford University) shows an ultrasound simulation that suggests that multiple scattering can be a dominant component in ultrasound. Because multiple scattering in ultrasound is ignored when multiple scattering is present it can significantly degrade image quality.
In the BEAM Lab, we are exploring model based and non-parametric approaches to suppressing multiple scattering based image degradation. Additionally, we aim to accomplish this while preserving the radio frequency signal that is crucial for advanced ultrasonic applications.
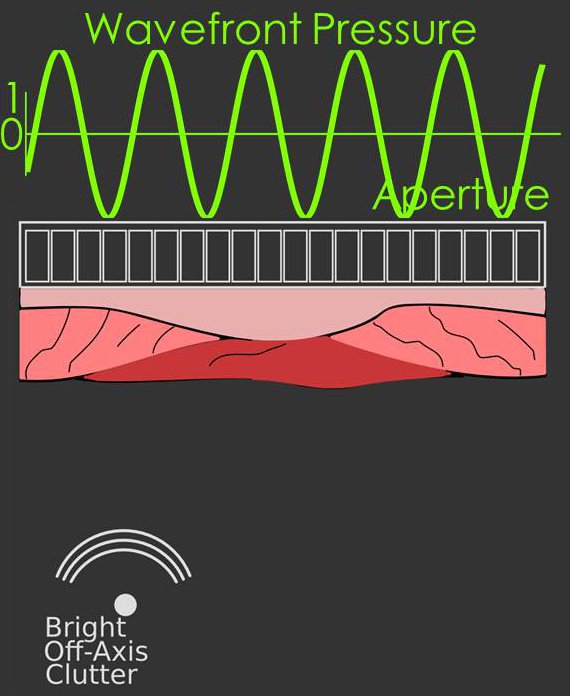
In classic beamforming the wavefronts arriving at the transducer can be well described by stationary
sinusoids e.g. cos(ωt) or sin(ωt) this includes waves originating far from the
transducer or waves originating close to the transducer after focusing.
 An example diagram of a stationary signal sampled by the array is shown in the figure on the right.
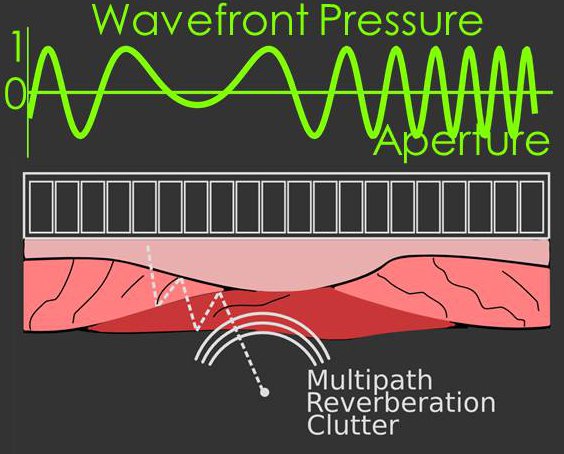
In addition to the stationary sinusoid model, we have shown wavefronts arriving after multiple
scattering (or misfocused wavefronts) can be modeled well by a non-stationary sinusoid,
i.e. a chirp signal. We have created a model that accounts for both the stationary and non-stationary
aspects of the signal, and when the model is applied to beamforming it suppresses wavefronts originating from
multiple scattering.
An example diagram of a stationary signal sampled by the array is shown in the figure on the right.
In addition to the stationary sinusoid model, we have shown wavefronts arriving after multiple
scattering (or misfocused wavefronts) can be modeled well by a non-stationary sinusoid,
i.e. a chirp signal. We have created a model that accounts for both the stationary and non-stationary
aspects of the signal, and when the model is applied to beamforming it suppresses wavefronts originating from
multiple scattering.
Example results are shown below for our model-based beamformer applied to in vivo human data.
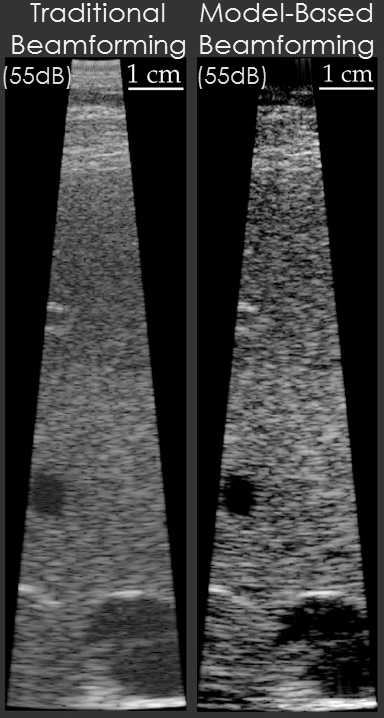
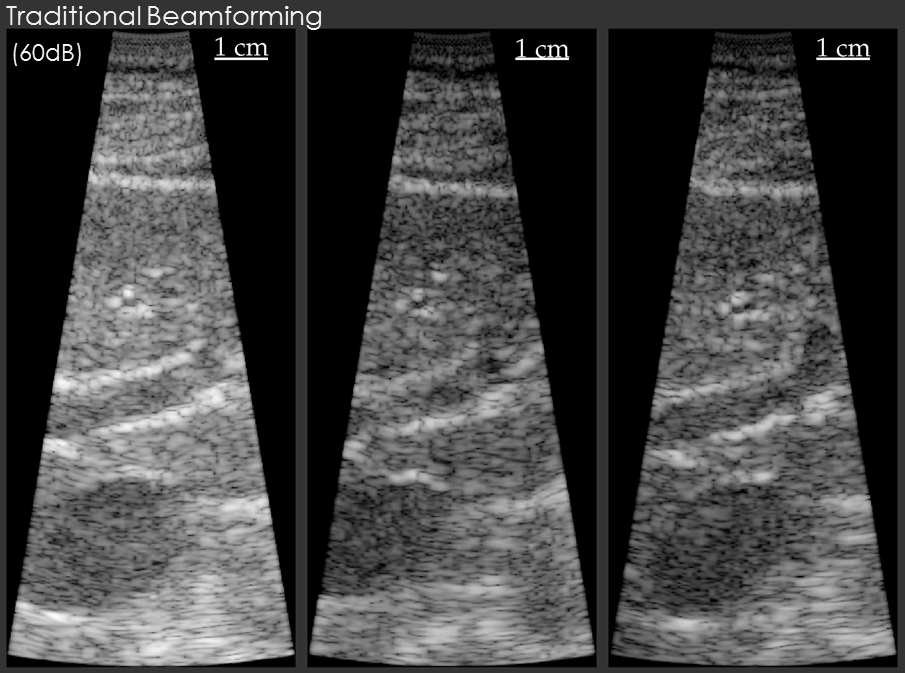
In the first example, the data beamformed using traditional methods has a "hazy" appearance that is common in in vivo imaging. The image beamformed using the model-based beamformer appear significantly sharper. The result is compelling, but a common criticism is that the same result could be obtained by changing the compression on the ultrasound scanner. To address this criticism another set of example data is shown. The second set of example data shows 3 images of the same structure in the same patient. When the data from the patient is beamformed using traditional methods the bile duct is not visible. When the data is beamformed using a model-based approach the bile duct is present in each case. The notable aspect of this example is that changing the system compression would not enable the visualization of the bile duct.
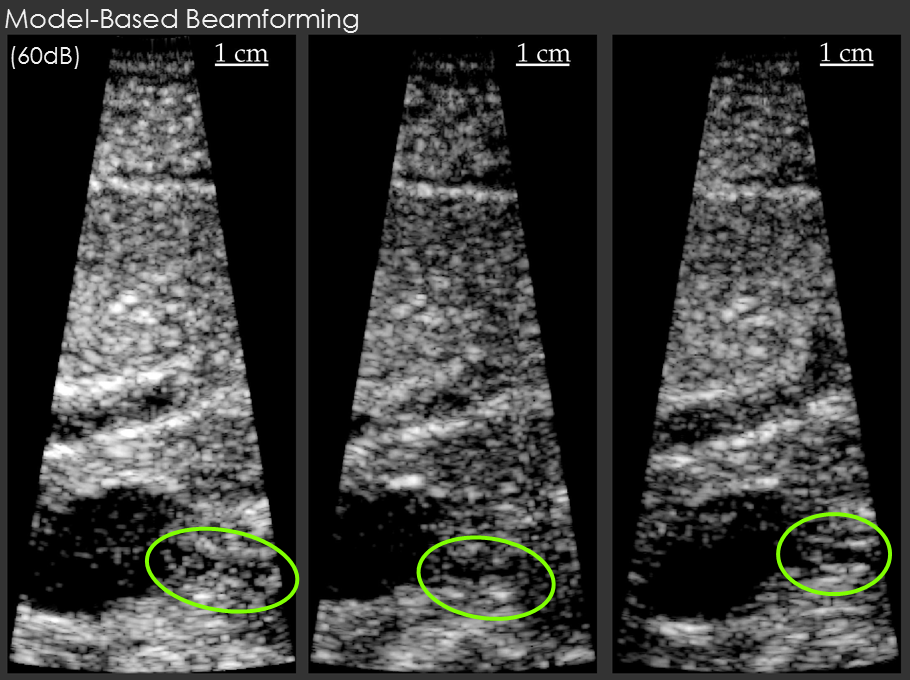
Note: Thanks to efforts by Chris Khan, the ADMIRE algorithm can be implemented in real-time, and that code is now available on our GitHub repository.
Beamforming with Deep Neural Networks
Our results above indicate that non-linear regression approaches to beamforming make a big difference.
Unfortunately, they're usually very slow and have some
other practical limitations, but there are other mathematical tools that can achieve similar
results much faster. These include deep neural networks. For our application, deep networks are attractive
because the training is slow, but once the networks are trained they run fast. We became the first
group to demonstrate how deep neural networks could be used for ultrasound beamforming to achieve state
of the art outcomes. Our early work is linked, and you can see more recent work under our publications:
click here.
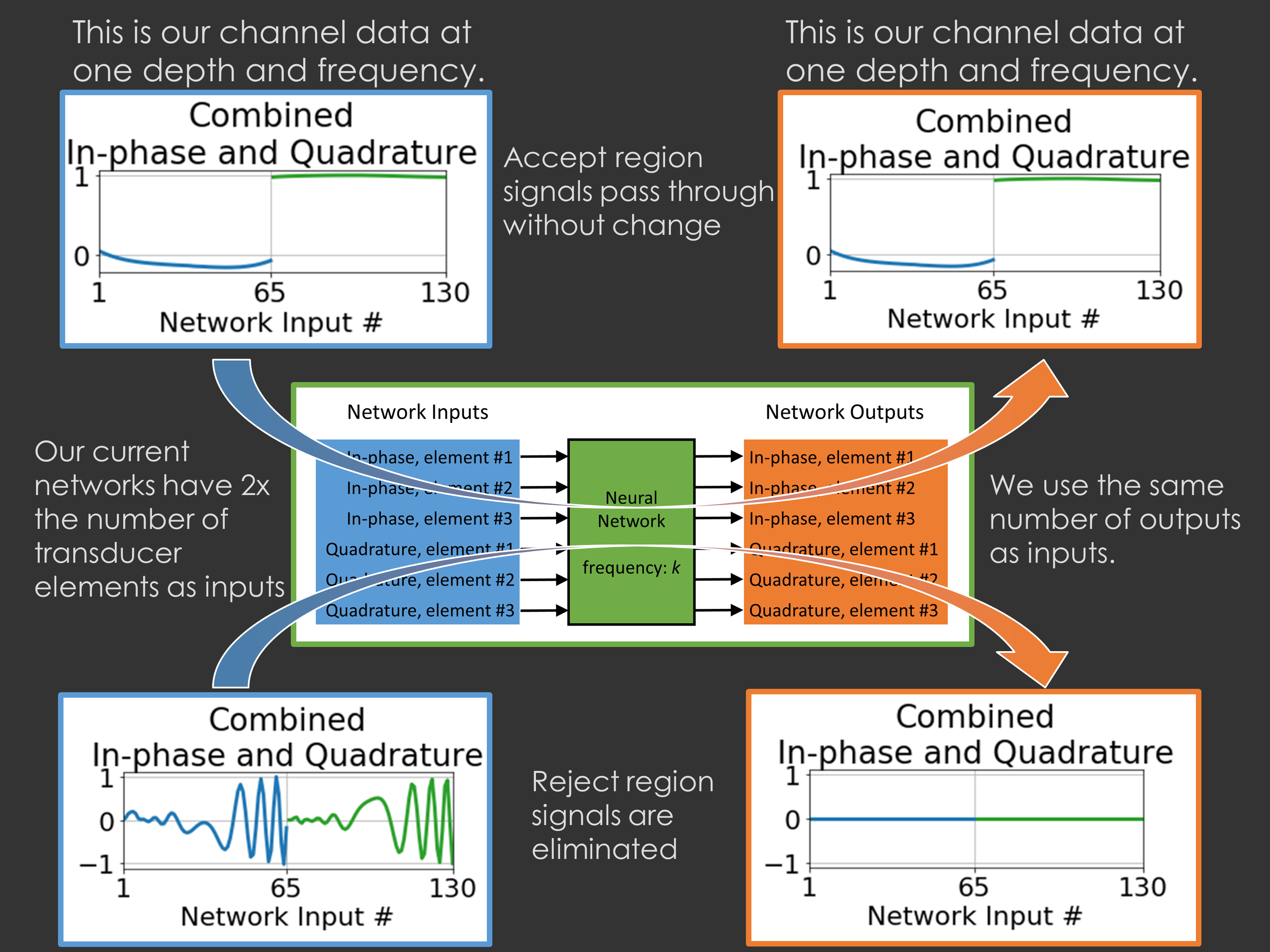
Our more recent work on deep networks has looked at how we can train deep networks with in vivo data even without having labeled training data: click here.
Brett Byram

Office: SC 5905
Phone: (615) 343-2327
b.byram _at_ vanderbilt.edu